User:Bci2
Projects
[edit]
Headline text
[edit]Inline linking of images: work in progress.
Images for DNA Structure Determination from X-Ray Patterns
[edit]File:ABDNAxrgpj.jpg
Acknowledgements:
- The author gratefully acknowledges the A- and B- DNA X-ray patterns reproduction rights in the US by Professor H.R. Wilson, F.R.S.
- The file "File:SLAC detector edit1.jpg" is credited on the Commons to User Mfield.
- The file "File:X ray diffraction.png" is credited on the Wikipedia Commons to author Thomas Splettstoesser.
- The file "File:Kappa goniometer animation.ogg" is credited to author Willow (Wikipedia User).
DNA Structures Derived from Analyses of X-ray Patterns
[edit]
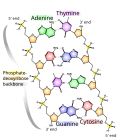
DNA exists in many possible conformations that include A-DNA, B-DNA, and Z-DNA forms, although, only B-DNA and Z-DNA have been directly observed in functional organisms. The conformation that DNA adopts depends on the hydration level, DNA sequence, the amount and direction of supercoiling, chemical modifications of the bases, the type and concentration of metal ions, as well as the presence of polyamines in solution.[2]
The first published reports of A-DNA X-ray diffraction patterns-- and also B-DNA-- employed analyses based on Patterson transforms[3][4] that provided only a limited amount of structural information for oriented fibers of DNA isolated from calf thymus. An alternate analysis was then proposed by Wilkins et al. in 1953[5] for B-DNA X-ray diffraction/scattering patterns of hydrated and oriented DNA fibers in terms of squares of Bessel functions. Watson and Crick then proposed to add DNA double-helix molecular modeling to the analysis.[6] of DNA X-ray diffraction patterns.
Although the `B-DNA form' is most common under the conditions found in cells,[7] it is not a well-defined conformation but a family or fuzzy set of DNA-conformations[8] that occur at the high hydration levels present in a wide variety of living cells. Their corresponding X-ray diffraction & scattering patterns are characteristic of molecular paracrystals with a significant degree of disorder (>20%)[9][10], and concomitantly the structure is not tractable using only the simplified standard Fourier transform/Bessel function analysis combined with the single DNA double-helix molecular model[6] of Crick and Watson. Such a standard analysis of DNA X-ray patterns, involving only Fourier transforms of Bessel functions[11] and DNA molecular models-- with several technical refinements[12]--is still in use for the analysis of A-DNA and Z-DNA X-ray diffraction patterns.
Compared to B-DNA, the A-DNA form is a wider right-handed spiral, with a shallow, wide minor groove and a narrower, deeper major groove. The A form occurs under non-physiological conditions in partially dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands, as well as in enzyme-DNA complexes.[13][14] Segments of DNA where the bases have been chemically modified by methylation may undergo a larger change in conformation and adopt the Z form. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form.[15] These unusual structures can be recognized by specific Z-DNA binding proteins and may be involved in the regulation of transcription.[16]
New section
[edit]Renowned Crystallographers
[edit]See links
[edit]- A- and B- DNA X-ray Patterns
- File:ABDNAxrgpj.jpg[17]
Notes
[edit]- ↑ Created from http://commons.wikimedia.org/wiki/File:ABDNAxrgpj.jpg
- ↑ Basu H, Feuerstein B, Zarling D, Shafer R, Marton L (1988). "Recognition of Z-RNA and Z-DNA determinants by polyamines in solution: experimental and theoretical studies". J Biomol Struct Dyn 6 (2): 299–309. PMID 2482766.
- ↑ Franklin, R.E. and Gosling, R.G. received 6 March 1953. Acta Cryst. (1953). 6, 673: The Structure of Sodium Thymonucleate Fibres I. The Influence of Water Content.; also Acta Cryst. 6, 678: The Structure of Sodium Thymonucleate Fibres II. The Cylindrically Symmetrical Patterson Function
- ↑ Franklin, Rosalind (1953). "Molecular Configuration in Sodium Thymonucleate. Franklin R. and Gosling R.G" (PDF). Nature 171: 740–741. DOI:10.1038/171740a0. PMID 13054694.
- ↑ Wilkins M.H.F., A.R. Stokes A.R. & Wilson, H.R. (1953). "Molecular Structure of Deoxypentose Nucleic Acids" (PDF). Nature 171: 738–740. DOI:10.1038/171738a0. PMID 13054693.
- ↑ a b Watson J.D. and Crick F.H.C. (1953). "A Structure for Deoxyribose Nucleic Acid" (PDF). Nature 171: 737–738. DOI:10.1038/171737a0. PMID 13054692.
- ↑ Leslie AG, Arnott S, Chandrasekaran R, Ratliff RL (1980). "Polymorphism of DNA double helices". J. Mol. Biol. 143 (1): 49–72. DOI:10.1016/0022-2836(80)90124-2. PMID 7441761.
- ↑ Baianu, I.C. (1980). "Structural Order and Partial Disorder in Biological systems". Bull. Math. Biol. 42 (4): 464–468. DOI:10.1016/0022-2836(80)90124-2.
- ↑ Hosemann R., Bagchi R.N., Direct analysis of diffraction by matter, North-Holland Publs., Amsterdam – New York, 1962
- ↑ Baianu I.C., X-ray scattering by partially disordered membrane systems, Acta Cryst. A, 34 (1978), 751–753.
- ↑ Bessel functions and diffraction by helical structures
- ↑ X-Ray Diffraction Patterns of Double-Helical Deoxyribonucleic Acid (DNA) Crystals
- ↑ Wahl M, Sundaralingam M (1997). "Crystal structures of A-DNA duplexes". Biopolymers 44 (1): 45–63. DOI:10.1002/(SICI)1097-0282(1997)44:1. PMID 9097733.
- ↑ Lu XJ, Shakked Z, Olson WK (2000). "A-form conformational motifs in ligand-bound DNA structures". J. Mol. Biol. 300 (4): 819–40. DOI:10.1006/jmbi.2000.3690. PMID 10891271.
- ↑ Rothenburg S, Koch-Nolte F, Haag F (2001). "DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles". Immunol Rev 184: 286–98. DOI:10.1034/j.1600-065x.2001.1840125.x. PMID 12086319.
- ↑ Oh D, Kim Y, Rich A (2002). "Z-DNA-binding proteins can act as potent effectors of gene expression in vivo". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16666–71. DOI:10.1073/pnas.262672699. PMID 12486233.
- ↑ http://commons.wikimedia.org/wiki/File:ABDNAxrgpj.jpg
New section: http://en.wikipedia.org/wiki/Molecular_models_of_DNA
[edit]http://en.wikipedia.org/wiki/Molecular_models_of_DNA
Molecular models of DNA structures are representations of the molecular geometry and topology of Deoxyribonucleic acid (DNA) molecules using one of several means, such as: closely packed spheres (CPK models) made of plastic, metal wires for 'skeletal models', graphic computations and animations by computers, artistic rendering, and so on, with the aim of simplifying and presenting the essential, physical and chemical, properties of DNA molecular structures either in vivo or in vitro. Altough DNA consists of relatively rigid, very large elongated biopolymer molecules called "fibers" or chains (that are made of repeating nucleotide units of four basic types, attached to deoxyribose and phospate groups), its molecular stucture in vivo undergoes dynamic configuration changes that involve dynamically attached water molecules and ions. Supercoiling, packing with histones in chromosome structures, and other such supramolecular aspects also involve in vivo DNA topology which is even more complex than DNA molecular geometry, thus turning molecular modeling of DNA into an especially challenging problem for both molecular biologists and biotechnologists. Like other large molecules and biopolymers, DNA often exists in multiple stable geometries (that is, it exhibits conformational isomerism) and configurational, quantum states which are close to each other in energy on the potential energy surface of the DNA molecule. Such geometries can also be computed, at least in principle, by employing ab initio quantum chemistry methods that have high accuracy for small molecules. Such quantum geometries define an important class of ab initio molecular models of DNA whose exploration has barely started. In an interesting twist of roles, the DNA molecule itself was proposed to be utilized for quantum computing.

The more advanced, computer-based molecular models of DNA involve molecular dynamics simulations as well as quantum mechanical computations of vibro-rotations, delocalized molecular orbitals (MOs), electric dipole moments, hydrogen-bonding, and so on.

Importance
[edit]From the very early stages of structural studies of DNA by X-ray diffraction and biochemical means, molecular models such as the Watson-Crick double-helix model were succesfully employed to solve the 'puzzle' of DNA structure, and also find how the latter relates to its key functions in living cells. The first high quality X-ray diffraction patterns of A-DNA were reported by Rosalind Franklin and Raymond Gosling in 1953[1]. The first calculations of the Fourier transform of an atomic helix were reported one year earlier by Cochran, Crick and Vand [2], and were followed in 1953 by the computation of the Fourier transform of a coiled-coil by Crick[3]. The first reports of a double-helix molecular model of B-DNA structure were made by Watson and Crick in 1953[4][5]. Last-but-not-least, Maurice F. Wilkins, A. Stokes and H.R. Wilson, reported the first X-ray patterns of in vivo B-DNA in partially oriented salmon sperm heads [6]. The development of the first correct double-helix molecular model of DNA by Crick and Watson may not have been possible without the biochemical evidence for the nucleotide base-pairing ([A---T]; [C---G]), or Chargaff's rules[7][8][9][10][11][12].
Examples of DNA molecular models
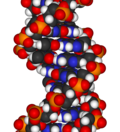
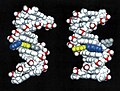
[edit]Animated molecular models allow one to visually explore the three-dimensional (3D) structure of DNA. The first DNA model is a space-filling, or CPK, model of the DNA double-helix whereas the third is an animated wire, or skeletal type, molecular model of DNA. The last two DNA molecular models in this series depict quadruplex DNA that may be involved in certain cancers[13][14]. The last figure on this panel is a molecular model of hydrogen bonds between water molecules in ice that are similar to those found in DNA.
- Spacefilling models or CPK models - a molecule is represented by overlapping spheres representing the atoms:
Images for DNA Structure Determination from X-Ray Patterns
[edit]The following images illustrate both the principles and the main steps involved in generating structural information from X-ray diffraction studies of oriented DNA fibers with the help of molecular models of DNA that are combined with crystallographic and mathematical analysis of the X-ray patterns.
Genomics and Biotechnology Applications of DNA molecular modeling
[edit]The following gallery of images illustrates various uses of DNA molecular modeling in Genomics and Biotechnology research applications from DNA repair to PCR and DNA nanostructures
X-ray diffraction
[edit]- NDB ID: UD0017 Database
- X-ray Atlas -database
- PDB files of coordinates for nucleic acid structures from X-ray diffraction by NA (incl. DNA) crystals
- Structure factors dowloadable files in CIF format
Neutron scattering
[edit]- ISIS neutron source
- ISIS pulsed neutron source:A world centre for science with neutrons & muons at Harwell, near Oxford, UK.
Electron microscopy
[edit]Spectroscopy
[edit]- Vibrational circular dichroism (VCD)
- FT-NMR[15][16]
- Microwave spectroscopy
- FT-IR
- FT-NIR
- Raman spectroscopy
- Fluorescence correlation spectroscopy and FRET[17][18][19].
Genomic and structural databases
[edit]- CBS Genome Atlas Database — contains examples of base skews.[20]
- The Z curve database of genomes — a 3-dimensional visualization and analysis tool of genomes[21].
- DNA and other nucleic acids' molecular models: Coordinate files of nucleic acids molecular structure models in PDB and CIF formats
Paracrystalline lattice models of B-DNA structures
[edit]A paracrystalline lattice, or paracrystal, is a molecular or atomic lattice with significant amounts (e.g., larger than a few percent) of partial disordering of molecular arranegements. Limiting cases of the paracrystal model are nanostructures, such as glasses, liquids, etc., that may possess only local ordering and no global order. Liquid crystals also have paracrystalline rather than crystalline structures.
Highly hydrated B-DNA occurs naturally in living cells in such a paracrystalline state, which is a dynamic one in spite of the relatively rigid DNA double-helix stabilized by parallel hydrogen bonds between the nucleotide base-pairs in the two complementary, helical DNA chains (see figures). For simplicity most DNA molecular models ommit both water and ions dynamically bound to B-DNA, and are thus less useful for understanding the dynamic behaviors of B-DNA in vivo. The physical and mathematical analysis of X-ray[22][23] and spectroscopic data for paracrystalline B-DNA is therefore much more complicated than that of crystalline, A-DNA X-ray diffraction patterns. The paracrystal model is also important for DNA technological applications such as DNA nanotechnology.
Notes
[edit]- ↑ Franklin, R.E. and Gosling, R.G. recd.6 March 1953. Acta Cryst. (1953). 6, 673 The Structure of Sodium Thymonucleate Fibres I. The Influence of Water Content Acta Cryst. (1953). and 6, 678 The Structure of Sodium Thymonucleate Fibres II. The Cylindrically Symmetrical Patterson Function.
- ↑ Cochran, W., Crick, F.H.C. and Vand V. 1952. The Structure of Synthetic Polypeptides. 1. The Transform of Atoms on a Helix. Acta Cryst. 5(5):581-586.
- ↑ Crick, F.H.C. 1953a. The Fourier Transform of a Coiled-Coil., Acta Crystallographica 6(8-9):685-689.
- ↑ Watson, J.D; Crick F.H.C. 1953a. Molecular Structure of Nucleic Acids- A Structure for Deoxyribose Nucleic Acid., Nature 171(4356):737-738.
- ↑ Watson, J.D; Crick F.H.C. 1953b. The Structure of DNA., Cold Spring Harbor Symposia on Qunatitative Biology 18:123-131.
- ↑ Wilkins M.H.F., A.R. Stokes A.R. & Wilson, H.R. (1953). "Molecular Structure of Deoxypentose Nucleic Acids" (PDF). Nature 171: 738–740. DOI:10.1038/171738a0. PMID 13054693.
- ↑ Elson D, Chargaff E (1952). "On the deoxyribonucleic acid content of sea urchin gametes". Experientia 8 (4): 143-145.
- ↑ Chargaff E, Lipshitz R, Green C (1952). "Composition of the deoxypentose nucleic acids of four genera of sea-urchin". J Biol Chem 195 (1): 155-160. PMID 14938364.
- ↑ Chargaff E, Lipshitz R, Green C, Hodes ME (1951). "The composition of the deoxyribonucleic acid of salmon sperm". J Biol Chem 192 (1): 223-230. PMID 14917668.
- ↑ Chargaff E (1951). "Some recent studies on the composition and structure of nucleic acids". J Cell Physiol Suppl 38 (Suppl).
- ↑ Magasanik B, Vischer E, Doniger R, Elson D, Chargaff E (1950). "The separation and estimation of ribonucleotides in minute quantities". J Biol Chem 186 (1): 37-50. PMID 14778802.
- ↑ Chargaff E (1950). "Chemical specificity of nucleic acids and mechanism of their enzymatic degradation". Experientia 6 (6): 201-209.
- ↑ http://www.phy.cam.ac.uk/research/bss/molbiophysics.php
- ↑ http://planetphysics.org/encyclopedia/TheoreticalBiophysics.html
- ↑ http://www.jonathanpmiller.com/Karplus.html- obtaining dihedral angles from 3J coupling constants
- ↑ http://www.spectroscopynow.com/FCKeditor/UserFiles/File/specNOW/HTML%20files/General_Karplus_Calculator.htm Another Javascript-like NMR coupling constant to dihedral
- ↑ FRET description
- ↑ doi:10.1016/S0959-440X(00)00190-1Recent advances in FRET: distance determination in protein–DNA complexes. Current Opinion in Structural Biology 2001, 11(2), 201-207
- ↑ http://www.fretimaging.org/mcnamaraintro.html FRET imaging introduction
- ↑ Hallin PF, David Ussery D (2004). "CBS Genome Atlas Database: A dynamic storage for bioinformatic results and DNA sequence data". Bioinformatics 20: 3682-3686.
- ↑ Zhang CT, Zhang R, Ou HY (2003). "The Z curve database: a graphic representation of genome sequences". Bioinformatics 19 (5): 593-599. doi:10.1093/bioinformatics/btg041
- ↑ Hosemann R., Bagchi R.N., Direct analysis of diffraction by matter, North-Holland Publs., Amsterdam – New York, 1962.
- ↑ Baianu, I.C. (1978). "X-ray scattering by partially disordered membrane systems.". Acta Cryst., A34 (5): 751–753. DOI:10.1107/S0567739478001540.
References
[edit]- F. Bessel, Untersuchung des Theils der planetarischen Störungen, Berlin Abhandlungen (1824), article 14.
- Sir Lawrence Bragg, FRS. The Crystalline State, A General survey. London: G. Bells and Sons, Ltd., vols. 1 and 2., 1966.
- Cantor, C. R. and Schimmel, P.R. Biophysical Chemistry, Parts I and II., San Franscisco: W.H. Freeman and Co. 1980. 1,800 pages.
- Voet, D. and J.G. Voet. Biochemistry, 2nd Edn., New York, Toronto, Singapore: John Wiley & Sons, Inc., 1995, ISBN: 0-471-58651-X., 1361 pages.
- Watson, G. N. A Treatise on the Theory of Bessel Functions., (1995) Cambridge University Press. ISBN 0-521-48391-3.
- Watson, James D. and Francis H.C. Crick. A structure for Deoxyribose Nucleic Acid (PDF). Nature 171, 737–738, 25 April 1953.
- Watson, James D. Molecular Biology of the Gene. New York and Amsterdam: W.A. Benjamin, Inc. 1965., 494 pages.
- Wentworth, W.E. Physical Chemistry. A short course., Malden (Mass.): Blackwell Science, Inc. 2000
- Herbert R. Wilson, FRS. Diffraction of X-rays by proteins, Nucleic Acids and Viruses., London: Edward Arnold (Publishers) Ltd. 1966.
- Kurt Wuthrich. NMR of Proteins and Nucleic Acids., New York, Brisbane,Chicester, Toronto, Singapore: J. Wiley & Sons. 1986., 292 pages.
See also
[edit]- DNA
- Molecular graphics
- DNA structure
- X-ray scattering
- Crystallography
- Crystal lattices
- Paracrystalline
- 2D-FT NMRI and Spectroscopy
- NMR Spectroscopy
- Molecular structure
- Molecular geometry
- Molecular topology
- DNA topology
- Nanostructure
- DNA nanotechnology
- Liquid crystal
- Glasses
- QMC@Home
- Sir Lawrence Bragg, FRS
- Sir John Randall
- James Watson
- Francis Crick
- Maurice Wilkins
- Herbert Wilson, FRS
- Alex Stokes
External links
[edit]- DNA the Double Helix Game From the official Nobel Prize web site
- Double Helix 1953–2003 National Centre for Biotechnology Education
- DNA under electron microscope
- The Secret Life of DNA - DNA Music compositions
- U.S. National DNA Day — watch videos and participate in real-time discusssions with scientists.
- Further details of mathematical and molecular analysis of DNA structure based on X-ray data
- Bessel functions corresponding to Fourier transforms of atomic or molecular helices.
Category:Molecular geometry Category:Helices Category:Diffraction Category:Molecular dynamics Category:Scattering Category:Quantum chemistry Category:Computational models Category:Molecular biology Category:Molecular genetics Category:Genomics Category:Genetics Category:Crystallography Category:Spectroscopy * Category:Polymers Category:Biomolecules Category:Nanotechnology Category:Biotechnology products Category:Classes of computers Category:Information theory
DNA Molecular Models
[edit]Molecular models of DNA structures are representations of the molecular geometry and topology of Deoxyribonucleic acid (DNA) molecules using one of several means, such as: closely packed spheres (CPK models) made of plastic, metal wires for 'skeletal models', graphic computations and animations by computers, artistic rendering, and so on, with the aim of simplifying and presenting the essential, physical and chemical, properties of DNA molecular structures either in vivo or in vitro. Computer molecular models also allow animations and molecular dynamics simulations that are very important for understanding how DNA functions in vivo. Thus, an old standing dynamic problem is how DNA "self-replication" takes place in living cells that should involve transient uncoiling of supercoiled DNA fibers. Altough DNA consists of relatively rigid, very large elongated biopolymer molecules called "fibers" or chains (that are made of repeating nucleotide units of four basic types, attached to deoxyribose and phospate groups), its molecular stucture in vivo undergoes dynamic configuration changes that involve dynamically attached water molecules and ions. Supercoiling, packing with histones in chromosome structures, and other such supramolecular aspects also involve in vivo DNA topology which is even more complex than DNA molecular geometry, thus turning molecular modeling of DNA into an especially challenging problem for both molecular biologists and biotechnologists. Like other large molecules and biopolymers, DNA often exists in multiple stable geometries (that is, it exhibits conformational isomerism) and configurational, quantum states which are close to each other in energy on the potential energy surface of the DNA molecule. Such geometries can also be computed, at least in principle, by employing ab initio quantum chemistry methods that have high accuracy for small molecules. Such quantum geometries define an important class of ab initio molecular models of DNA whose exploration has barely started.

In an interesting twist of roles, the DNA molecule itself was proposed to be utilized for quantum computing. Both DNA nanostructures as well as DNA 'computing' biochips have been built (see biochip image at right).
The more advanced, computer-based molecular models of DNA involve molecular dynamics simulations as well as quantum mechanical computations of vibro-rotations, delocalized molecular orbitals (MOs), electric dipole moments, hydrogen-bonding, and so on.

Importance
[edit]From the very early stages of structural studies of DNA by X-ray diffraction and biochemical means, molecular models such as the Watson-Crick double-helix model were succesfully employed to solve the 'puzzle' of DNA structure, and also find how the latter relates to its key functions in living cells. The first high quality X-ray diffraction patterns of A-DNA were reported by Rosalind Franklin and Raymond Gosling in 1953[1]. The first calculations of the Fourier transform of an atomic helix were reported one year earlier by Cochran, Crick and Vand [2], and were followed in 1953 by the computation of the Fourier transform of a coiled-coil by Crick[3]. The first reports of a double-helix molecular model of B-DNA structure were made by Watson and Crick in 1953[4][5]. Last-but-not-least, Maurice F. Wilkins, A. Stokes and H.R. Wilson, reported the first X-ray patterns of in vivo B-DNA in partially oriented salmon sperm heads [6]. The development of the first correct double-helix molecular model of DNA by Crick and Watson may not have been possible without the biochemical evidence for the nucleotide base-pairing ([A---T]; [C---G]), or Chargaff's rules[7][8][9][10][11][12].
Examples of DNA molecular models
[edit]Animated molecular models allow one to visually explore the three-dimensional (3D) structure of DNA. The first DNA model is a space-filling, or CPK, model of the DNA double-helix whereas the third is an animated wire, or skeletal type, molecular model of DNA. The last two DNA molecular models in this series depict quadruplex DNA that may be involved in certain cancers[13][14]. The last figure on this panel is a molecular model of hydrogen bonds between water molecules in ice that are similar to those found in DNA.
- Spacefilling models or CPK models - a molecule is represented by overlapping spheres representing the atoms:
Images for DNA Structure Determination from X-Ray Patterns
[edit]The following images illustrate both the principles and the main steps involved in generating structural information from X-ray diffraction studies of oriented DNA fibers with the help of molecular models of DNA that are combined with crystallographic and mathematical analysis of the X-ray patterns. From left to right the gallery of images shows:
- First row:
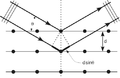
- 1. Constructive X-ray interference, or diffraction, following Bragg's Law of X-ray "reflection by the crystal planes";
- 2. A comparison of A-DNA (crystalline) and highly hydrated B-DNA (paracrystalline) X-ray diffraction, and respectively, X-ray scattering patterns (courtesy of Dr. Herbert R. Wilson, FRS- see refs. list);
- 3. Purified DNA precipitated in a water jug;
- 4. The major steps involved in DNA structure determination by X-ray crystallography showing the important role played by molecular models of DNA structure in this iterative, structure--determination process;
- Second row:
- 5. Photo of a modern X-ray diffractometer employed for recording X-ray patterns of DNA with major components: X-ray source, goniometer, sample holder, X-ray detector and/or plate holder;
- 6. Illustrated animation of an X-ray goniometer;
- 7. X-ray detector at the SLAC synchrotron facility;
- 8. Neutron scattering facility at ISIS in UK;
- Third and fourth rows: Molecular models of DNA structure at various scales; figure #11 is an actual electron micrograph of a DNA fiber bundle, presumably of a single bacterial chromosome loop.
Paracrystalline lattice models of B-DNA structures
[edit]A paracrystalline lattice, or paracrystal, is a molecular or atomic lattice with significant amounts (e.g., larger than a few percent) of partial disordering of molecular arranegements. Limiting cases of the paracrystal model are nanostructures, such as glasses, liquids, etc., that may possess only local ordering and no global order. Liquid crystals also have paracrystalline rather than crystalline structures.
Highly hydrated B-DNA occurs naturally in living cells in such a paracrystalline state, which is a dynamic one in spite of the relatively rigid DNA double-helix stabilized by parallel hydrogen bonds between the nucleotide base-pairs in the two complementary, helical DNA chains (see figures). For simplicity most DNA molecular models ommit both water and ions dynamically bound to B-DNA, and are thus less useful for understanding the dynamic behaviors of B-DNA in vivo. The physical and mathematical analysis of X-ray[15][16] and spectroscopic data for paracrystalline B-DNA is therefore much more complicated than that of crystalline, A-DNA X-ray diffraction patterns. The paracrystal model is also important for DNA technological applications such as DNA nanotechnology. Novel techniques that combine X-ray diffraction of DNA with X-ray microscopy in hydrated living cells are now also being developed (see, for example, "Application of X-ray microscopy in the analysis of living hydrated cells").
Genomic and Biotechnology Applications of DNA molecular modeling
[edit]The following gallery of images illustrates various uses of DNA molecular modeling in Genomics and Biotechnology research applications from DNA repair to PCR and DNA nanostructures; each slide contains its own explanation and/or details. The first slide presents an overview of DNA applications, including DNA molecular models, with emphasis on Genomics and Biotechnology.
Gallery: DNA Molecular modeling applications
[edit]X-ray diffraction
[edit]- NDB ID: UD0017 Database
- X-ray Atlas -database
- PDB files of coordinates for nucleic acid structures from X-ray diffraction by NA (incl. DNA) crystals
- Structure factors dowloadable files in CIF format
Neutron scattering
[edit]- ISIS neutron source
- ISIS pulsed neutron source:A world centre for science with neutrons & muons at Harwell, near Oxford, UK.
X-ray microscopy
[edit]Electron microscopy
[edit]Atomic Force Microscopy (AFM)
[edit]Two-dimensional DNA junction arrays have been visualized by Atomic Force Microscopy (AFM)[17]. Other imaging resources for AFM/Scanning probe microscopy(SPM) can be freely accessed at:
Gallery of AFM Images
[edit]Mass spectrometry--Maldi informatics
[edit]Spectroscopy
[edit]- Vibrational circular dichroism (VCD)
- FT-NMR[18][19]
- NMR microscopy[20]
- Microwave spectroscopy
- FT-IR
- FT-NIR[21][22][23]
- Spectral, Hyperspectral, and Chemical imaging)[24][25][26][27][28][29][30].
- Raman spectroscopy/microscopy[31] and CARS[32].
- Fluorescence correlation spectroscopy[33][34][35][36][37][38][39][40], Fluorescence cross-correlation spectroscopy and FRET[41][42][43].
- Confocal microscopy[44]
Gallery: CARS (Raman spectroscopy), Fluorescence confocal microscopy, and Hyperspectral imaging
[edit]Genomic and structural databases
[edit]- CBS Genome Atlas Database — contains examples of base skews.[45]
- The Z curve database of genomes — a 3-dimensional visualization and analysis tool of genomes[46].
- DNA and other nucleic acids' molecular models: Coordinate files of nucleic acids molecular structure models in PDB and CIF formats
Notes
[edit]- ↑ Franklin, R.E. and Gosling, R.G. recd.6 March 1953. Acta Cryst. (1953). 6, 673 The Structure of Sodium Thymonucleate Fibres I. The Influence of Water Content Acta Cryst. (1953). and 6, 678 The Structure of Sodium Thymonucleate Fibres II. The Cylindrically Symmetrical Patterson Function.
- ↑ Cochran, W., Crick, F.H.C. and Vand V. 1952. The Structure of Synthetic Polypeptides. 1. The Transform of Atoms on a Helix. Acta Cryst. 5(5):581-586.
- ↑ Crick, F.H.C. 1953a. The Fourier Transform of a Coiled-Coil., Acta Crystallographica 6(8-9):685-689.
- ↑ Watson, J.D; Crick F.H.C. 1953a. Molecular Structure of Nucleic Acids- A Structure for Deoxyribose Nucleic Acid., Nature 171(4356):737-738.
- ↑ Watson, J.D; Crick F.H.C. 1953b. The Structure of DNA., Cold Spring Harbor Symposia on Qunatitative Biology 18:123-131.
- ↑ Wilkins M.H.F., A.R. Stokes A.R. & Wilson, H.R. (1953). "Molecular Structure of Deoxypentose Nucleic Acids" (PDF). Nature 171: 738–740. DOI:10.1038/171738a0. PMID 13054693.
- ↑ Elson D, Chargaff E (1952). "On the deoxyribonucleic acid content of sea urchin gametes". Experientia 8 (4): 143-145.
- ↑ Chargaff E, Lipshitz R, Green C (1952). "Composition of the deoxypentose nucleic acids of four genera of sea-urchin". J Biol Chem 195 (1): 155-160. PMID 14938364.
- ↑ Chargaff E, Lipshitz R, Green C, Hodes ME (1951). "The composition of the deoxyribonucleic acid of salmon sperm". J Biol Chem 192 (1): 223-230. PMID 14917668.
- ↑ Chargaff E (1951). "Some recent studies on the composition and structure of nucleic acids". J Cell Physiol Suppl 38 (Suppl).
- ↑ Magasanik B, Vischer E, Doniger R, Elson D, Chargaff E (1950). "The separation and estimation of ribonucleotides in minute quantities". J Biol Chem 186 (1): 37-50. PMID 14778802.
- ↑ Chargaff E (1950). "Chemical specificity of nucleic acids and mechanism of their enzymatic degradation". Experientia 6 (6): 201-209.
- ↑ http://www.phy.cam.ac.uk/research/bss/molbiophysics.php
- ↑ http://planetphysics.org/encyclopedia/TheoreticalBiophysics.html
- ↑ Hosemann R., Bagchi R.N., Direct analysis of diffraction by matter, North-Holland Publs., Amsterdam – New York, 1962.
- ↑ Baianu, I.C. (1978). "X-ray scattering by partially disordered membrane systems.". Acta Cryst., A34 (5): 751–753. DOI:10.1107/S0567739478001540.
- ↑ Mao, Chengde; Sun, Weiqiong & Seeman, Nadrian C. (16 June 1999). "Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy". Journal of the American Chemical Society 121 (23): 5437–5443. DOI:10.1021/ja9900398. ISSN 0002-7863.
- ↑ http://www.jonathanpmiller.com/Karplus.html- obtaining dihedral angles from 3J coupling constants
- ↑ http://www.spectroscopynow.com/FCKeditor/UserFiles/File/specNOW/HTML%20files/General_Karplus_Calculator.htm Another Javascript-like NMR coupling constant to dihedral
- ↑ Lee, S. C. et al., (2001). One Micrometer Resolution NMR Microscopy. J. Magn. Res., 150: 207-213.
- ↑ Near Infrared Microspectroscopy, Fluorescence Microspectroscopy,Infrared Chemical Imaging and High Resolution Nuclear Magnetic Resonance Analysis of Soybean Seeds, Somatic Embryos and Single Cells., Baianu, I.C. et al. 2004., In Oil Extraction and Analysis., D. Luthria, Editor pp.241-273, AOCS Press., Champaign, IL.
- ↑ Single Cancer Cell Detection by Near Infrared Microspectroscopy, Infrared Chemical Imaging and Fluorescence Microspectroscopy.2004.I. C. Baianu, D. Costescu, N. E. Hofmann and S. S. Korban, q-bio/0407006 (July 2004)
- ↑ Raghavachari, R., Editor. 2001. Near-Infrared Applications in Biotechnology, Marcel-Dekker, New York, NY.
- ↑ http://www.imaging.net/chemical-imaging/ Chemical imaging
- ↑ http://www.malvern.com/LabEng/products/sdi/bibliography/sdi_bibliography.htm E. N. Lewis, E. Lee and L. H. Kidder, Combining Imaging and Spectroscopy: Solving Problems with Near-Infrared Chemical Imaging. Microscopy Today, Volume 12, No. 6, 11/2004.
- ↑ D.S. Mantus and G. H. Morrison. 1991. Chemical imaging in biology and medicine using ion microscopy., Microchimica Acta, 104, (1-6) January 1991, doi: 10.1007/BF01245536
- ↑ Near Infrared Microspectroscopy, Fluorescence Microspectroscopy,Infrared Chemical Imaging and High Resolution Nuclear Magnetic Resonance Analysis of Soybean Seeds, Somatic Embryos and Single Cells., Baianu, I.C. et al. 2004., In Oil Extraction and Analysis., D. Luthria, Editor pp.241-273, AOCS Press., Champaign, IL.
- ↑ Single Cancer Cell Detection by Near Infrared Microspectroscopy, Infrared Chemical Imaging and Fluorescence Microspectroscopy.2004.I. C. Baianu, D. Costescu, N. E. Hofmann and S. S. Korban, q-bio/0407006 (July 2004)
- ↑ J. Dubois, G. Sando, E. N. Lewis, Near-Infrared Chemical Imaging, A Valuable Tool for the Pharmaceutical Industry, G.I.T. Laboratory Journal Europe, No.1-2, 2007.
- ↑ Applications of Novel Techniques to Health Foods, Medical and Agricultural Biotechnology.(June 2004).,I. C. Baianu, P. R. Lozano, V. I. Prisecaru and H. C. Lin q-bio/0406047
- ↑ Chemical Imaging Without Dyeing
- ↑ C.L. Evans and X.S. Xie.2008. Coherent Anti-Stokes Raman Scattering Microscopy: Chemical Imaging for Biology and Medicine., doi:10.1146/annurev.anchem.1.031207.112754 Annual Review of Analytical Chemistry, 1: 883-909.
- ↑ Eigen, M., Rigler, M. Sorting single molecules: application to diagnostics and evolutionary biotechnology,(1994) Proc. Natl. Acad. Sci. USA, 91,5740-5747.
- ↑ Rigler, M. Fluorescence correlations, single molecule detection and large number screening. Applications in biotechnology,(1995) J. Biotechnol., 41,177-186.
- ↑ Rigler R. and Widengren J. (1990). Ultrasensitive detection of single molecules by fluorescence correlation spectroscopy, BioScience (Ed. Klinge & Owman) p.180.
- ↑ Single Cancer Cell Detection by Near Infrared Microspectroscopy, Infrared Chemical Imaging and Fluorescence Microspectroscopy.2004.I. C. Baianu, D. Costescu, N. E. Hofmann, S. S. Korban and et al., q-bio/0407006 (July 2004)
- ↑ Oehlenschläger F., Schwille P. and Eigen M. (1996). Detection of HIV-1 RNA by nucleic acid sequence-based amplification combined with fluorescence correlation spectroscopy, Proc. Natl. Acad. Sci. USA 93:1281.
- ↑ Bagatolli, L.A., and Gratton, E. (2000). Two-photon fluorescence microscopy of coexisting lipid domains in giant unilamellar vesicles of binary phospholipid mixtures. Biophys J., 78:290-305.
- ↑ Schwille, P., Haupts, U., Maiti, S., and Webb. W.(1999). Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophysical Journal, 77(10):2251-2265.
- ↑ Near Infrared Microspectroscopy, Fluorescence Microspectroscopy,Infrared Chemical Imaging and High Resolution Nuclear Magnetic Resonance Analysis of Soybean Seeds, Somatic Embryos and Single Cells., Baianu, I.C. et al. 2004., In Oil Extraction and Analysis., D. Luthria, Editor pp.241-273, AOCS Press., Champaign, IL.
- ↑ FRET description
- ↑ doi:10.1016/S0959-440X(00)00190-1Recent advances in FRET: distance determination in protein–DNA complexes. Current Opinion in Structural Biology 2001, 11(2), 201-207
- ↑ http://www.fretimaging.org/mcnamaraintro.html FRET imaging introduction
- ↑ Eigen, M., and Rigler, R. (1994). Sorting single molecules: Applications to diagnostics and evolutionary biotechnology, Proc. Natl. Acad. Sci. USA 91:5740.
- ↑ Hallin PF, David Ussery D (2004). "CBS Genome Atlas Database: A dynamic storage for bioinformatic results and DNA sequence data". Bioinformatics 20: 3682-3686.
- ↑ Zhang CT, Zhang R, Ou HY (2003). "The Z curve database: a graphic representation of genome sequences". Bioinformatics 19 (5): 593-599. doi:10.1093/bioinformatics/btg041
References
[edit]- Applications of Novel Techniques to Health Foods, Medical and Agricultural Biotechnology.(June 2004) I. C. Baianu, P. R. Lozano, V. I. Prisecaru and H. C. Lin., q-bio/0406047.
- F. Bessel, Untersuchung des Theils der planetarischen Störungen, Berlin Abhandlungen (1824), article 14.
- Sir Lawrence Bragg, FRS. The Crystalline State, A General survey. London: G. Bells and Sons, Ltd., vols. 1 and 2., 1966., 2024 pages.
- Cantor, C. R. and Schimmel, P.R. Biophysical Chemistry, Parts I and II., San Franscisco: W.H. Freeman and Co. 1980. 1,800 pages.
- Eigen, M., and Rigler, R. (1994). Sorting single molecules: Applications to diagnostics and evolutionary biotechnology, Proc. Natl. Acad. Sci. USA 91:5740.
- Raghavachari, R., Editor. 2001. Near-Infrared Applications in Biotechnology, Marcel-Dekker, New York, NY.
- Rigler R. and Widengren J. (1990). Ultrasensitive detection of single molecules by fluorescence correlation spectroscopy, BioScience (Ed. Klinge & Owman) p.180.
- Single Cancer Cell Detection by Near Infrared Microspectroscopy, Infrared Chemical Imaging and Fluorescence Microspectroscopy.2004. I. C. Baianu, D. Costescu, N. E. Hofmann, S. S. Korban and et al., q-bio/0407006 (July 2004).
- Voet, D. and J.G. Voet. Biochemistry, 2nd Edn., New York, Toronto, Singapore: John Wiley & Sons, Inc., 1995, ISBN: 0-471-58651-X., 1361 pages.
- Watson, G. N. A Treatise on the Theory of Bessel Functions., (1995) Cambridge University Press. ISBN 0-521-48391-3.
- Watson, James D. and Francis H.C. Crick. A structure for Deoxyribose Nucleic Acid (PDF). Nature 171, 737–738, 25 April 1953.
- Watson, James D. Molecular Biology of the Gene. New York and Amsterdam: W.A. Benjamin, Inc. 1965., 494 pages.
- Wentworth, W.E. Physical Chemistry. A short course., Malden (Mass.): Blackwell Science, Inc. 2000.
- Herbert R. Wilson, FRS. Diffraction of X-rays by proteins, Nucleic Acids and Viruses., London: Edward Arnold (Publishers) Ltd. 1966.
- Kurt Wuthrich. NMR of Proteins and Nucleic Acids., New York, Brisbane,Chicester, Toronto, Singapore: J. Wiley & Sons. 1986., 292 pages.
- Robinson, Bruche H.; Seeman, Nadrian C. (August 1987). "The Design of a Biochip: A Self-Assembling Molecular-Scale Memory Device". Protein Engineering 1 (4): 295–300. ISSN 0269-2139. Link
- Rothemund, Paul W. K.; Ekani-Nkodo, Axel; Papadakis, Nick; Kumar, Ashish; Fygenson, Deborah Kuchnir & Winfree, Erik (22 December 2004). "Design and Characterization of Programmable DNA Nanotubes". Journal of the American Chemical Society 126 (50): 16344–16352. DOI:10.1021/ja044319l. ISSN 0002-7863.
- Keren, K.; Kinneret Keren, Rotem S. Berman, Evgeny Buchstab, Uri Sivan, Erez Braun (November 2003). "DNA-Templated Carbon Nanotube Field-Effect Transistor". Science 302 (6549): 1380–1382. DOI:10.1126/science.1091022. ISSN 1095-9203.
- Zheng, Jiwen; Constantinou, Pamela E.; Micheel, Christine; Alivisatos, A. Paul; Kiehl, Richard A. & Seeman Nadrian C. (2006). "2D Nanoparticle Arrays Show the Organizational Power of Robust DNA Motifs". Nano Letters 6: 1502–1504. DOI:10.1021/nl060994c. ISSN 1530-6984.
- Cohen, Justin D.; Sadowski, John P.; Dervan, Peter B. (2007). "Addressing Single Molecules on DNA Nanostructures". Angewandte Chemie 46 (42): 7956–7959. DOI:10.1002/anie.200702767. ISSN 0570-0833.
- Mao, Chengde; Sun, Weiqiong & Seeman, Nadrian C. (16 June 1999). "Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy". Journal of the American Chemical Society 121 (23): 5437–5443. DOI:10.1021/ja9900398. ISSN 0002-7863.
- Constantinou, Pamela E.; Wang, Tong; Kopatsch, Jens; Israel, Lisa B.; Zhang, Xiaoping; Ding, Baoquan; Sherman, William B.; Wang, Xing; Zheng, Jianping; Sha, Ruojie & Seeman, Nadrian C. (2006). "Double cohesion in structural DNA nanotechnology". Organic and Biomolecular Chemistry 4: 3414–3419. DOI:10.1039/b605212f.
See also
[edit]- DNA
- Molecular graphics
- DNA structure
- X-ray scattering
- Neutron scattering
- Crystallography
- Crystal lattices
- Paracrystalline lattices/Paracrystals
- 2D-FT NMRI and Spectroscopy
- NMR Spectroscopy
- Microwave spectroscopy
- Two-dimensional IR spectroscopy
- Spectral imaging
- Hyperspectral imaging
- Chemical imaging
- NMR microscopy
- VCD or Vibrational circular dichroism
- FRET and FCS- Fluorescence correlation spectroscopy
- Fluorescence cross-correlation spectroscopy (FCCS)
- Molecular structure
- Molecular geometry
- Molecular topology
- DNA topology
- Nanostructure
- DNA nanotechnology
- Imaging
- Atomic force microscopy
- X-ray microscopy
- Liquid crystal
- Glasses
- QMC@Home
- Sir Lawrence Bragg, FRS
- Sir John Randall
- James Watson
- Francis Crick
- Maurice Wilkins
- Herbert Wilson, FRS
- Alex Stokes
External links
[edit]- DNA the Double Helix Game From the official Nobel Prize web site
- MDDNA: Structural Bioinformatics of DNA
- Double Helix 1953–2003 National Centre for Biotechnology Education
- DNA under electron microscope
- Ascalaph DNA — Commercial software for DNA modeling
- DNAlive: a web interface to compute DNA physical properties. Also allows cross-linking of the results with the UCSC Genome browser and DNA dynamics.
- DiProDB: Dinucleotide Property Database. The database is designed to collect and analyse thermodynamic, structural and other dinucleotide properties.
- Further details of mathematical and molecular analysis of DNA structure based on X-ray data
- Bessel functions corresponding to Fourier transforms of atomic or molecular helices.
- Application of X-ray microscopy in analysis of living hydrated cells
- Characterization in nanotechnology some pdfs
- overview of STM/AFM/SNOM principles with educative videos
- SPM Image Gallery - AFM STM SEM MFM NSOM and More
- How SPM Works
- U.S. National DNA Day — watch videos and participate in real-time discusssions with scientists.
- The Secret Life of DNA - DNA Music compositions
Template:Nucleic acids Template:SPM2
Category:Molecular geometry Category:Helices Category:Diffraction Category:Molecular dynamics Category:Scattering Category:Quantum chemistry Category:Computational models Category:Molecular biology Category:Molecular genetics Category:Genomics Category:Genetics Category:Crystallography Category:Spectroscopy * Category:Polymers Category:Biomolecules Category:Nanotechnology Category:Biotechnology products Category:Databases Category:Imaging Category:Microscopic images Category:Scanning probe microscopy Category:Fluorescence Category:Database management systems Category:Classes of computers Category:Information theory Category:Nobel laureates in Physiology or Medicine Category:Molecular biologists Category:Biophysicists
See also
[edit]American-Romanian Academy of Arts and Sciences American Romanian Academy of Arts and Sciences