File:41598 2017 2668 Fig2 HTML.webp
From Wikimedia Commons, the free media repository
Jump to navigation
Jump to search
41598_2017_2668_Fig2_HTML.webp (675 × 587 pixels, file size: 80 KB, MIME type: image/webp)
File information
Structured data
Captions
Captions
Add a one-line explanation of what this file represents
Summary[edit]
| Description41598 2017 2668 Fig2 HTML.webp |
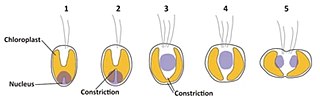
English: Identification of the nucleus-encoded nucleomorph-targeted histone H2A in Guillardia theta. (a) Alignment of the N-terminal sequences of histone H2A proteins. The N-terminal of the G. theta histone H2A (GuiTh_1001763) is composed of a signal peptide and a transit peptide-like sequence. No signal peptide is predicted in other G. theta nucleus-encoded histone H2A (GuiTh_44948, GuiTh_49238 and GuiTh_47177) or histone H2A variants (GuiTh_98800 and GuiTh_96636). Histone H2A sequences of the haptophyte Chrysochromulina sp. CCMP2919 (GI#922855917), the haptophyte Emiliania huxleyi (GI#551564196), and the red alga Cyanidioschyzon merolae (GI#449017866 and GI#449019224) were also used for the alignment. The asterisk indicates an amino acid that is identical in all sequences and the conserved region is boxed in yellow. (b) Phylogenetic tree of histone H2A inferred from 105 amino acid sequences. The histone H2A sequences of G. theta are indicated in bold and boxed in gray. H2A variants were used as the outgroup. The bootstrap value and Bayesian posterior probability are shown at the node when the values exceed 50% (bootstrap) or 0.80 (posterior probability). The JGI accession number or GI number is shown on the right side of the species name. (c) Schematic drawing of the classification of the cell cycle stages based on the localization of the nucleus and the shape of the chloroplast. (d) DAPI-staining images showing the representative cells of the stages 1 to 5. DIC, images of differential interference contact; Chl, chloroplast autofluorescence; Chl/DAPI, merged images of Chl and DAPI. The double arrowhead indicates constriction of the chloroplast division site. Scale bar = 5 µm. (e) Immunofluorescent micrographs showing the localization of the nucleus-encoded nucleomorph H2A. DIC, images of differential interference contact; Chl, chloroplast autofluorescence; Anti-Nm H2A, nucleomorph H2A detected with the antibody; DAPI, DNA stained with DAPI; H2A/DAPI, merged images of H2A and DAPI. The arrow and arrowhead indicate DAPI signal of nucleus and nucleomorph, respectively. Scale bar = 5 µm. |
||
| Date | |||
| Source |
Fig. 2 at https://www.nature.com/articles/s41598-017-02668-2 Regulation of chloroplast and nucleomorph replication by the cell cycle in the cryptophyte Guillardia theta. In: Scientific Reports volume 7, Article number: 2345. doi:10.1038/s41598-017-02668-2 |
||
| Author | Ryo Onuma, Neha Mishra, Shin-ya Miyagishima | ||
| Other versions |
|
Licensing[edit]
This file is licensed under the Creative Commons Attribution-Share Alike 4.0 International license.
- You are free:
- to share – to copy, distribute and transmit the work
- to remix – to adapt the work
- Under the following conditions:
- attribution – You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- share alike – If you remix, transform, or build upon the material, you must distribute your contributions under the same or compatible license as the original.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 14:23, 19 August 2021 |  | 675 × 587 (80 KB) | Ernsts (talk | contribs) | Uploaded a work by Ryo Onuma, Neha Mishra, Shin-ya Miyagishima from Fig. 2 at https://www.nature.com/articles/s41598-017-02668-2 Regulation of chloroplast and nucleomorph replication by the cell cycle in the cryptophyte ''Guillardia theta''. In: Scientific Reports volume 7, Article number: 2345. doi:10.1038/s41598-017-02668-2 50px|class=noviewer with UploadWizard |
You cannot overwrite this file.
File usage on Commons
The following 2 pages use this file: